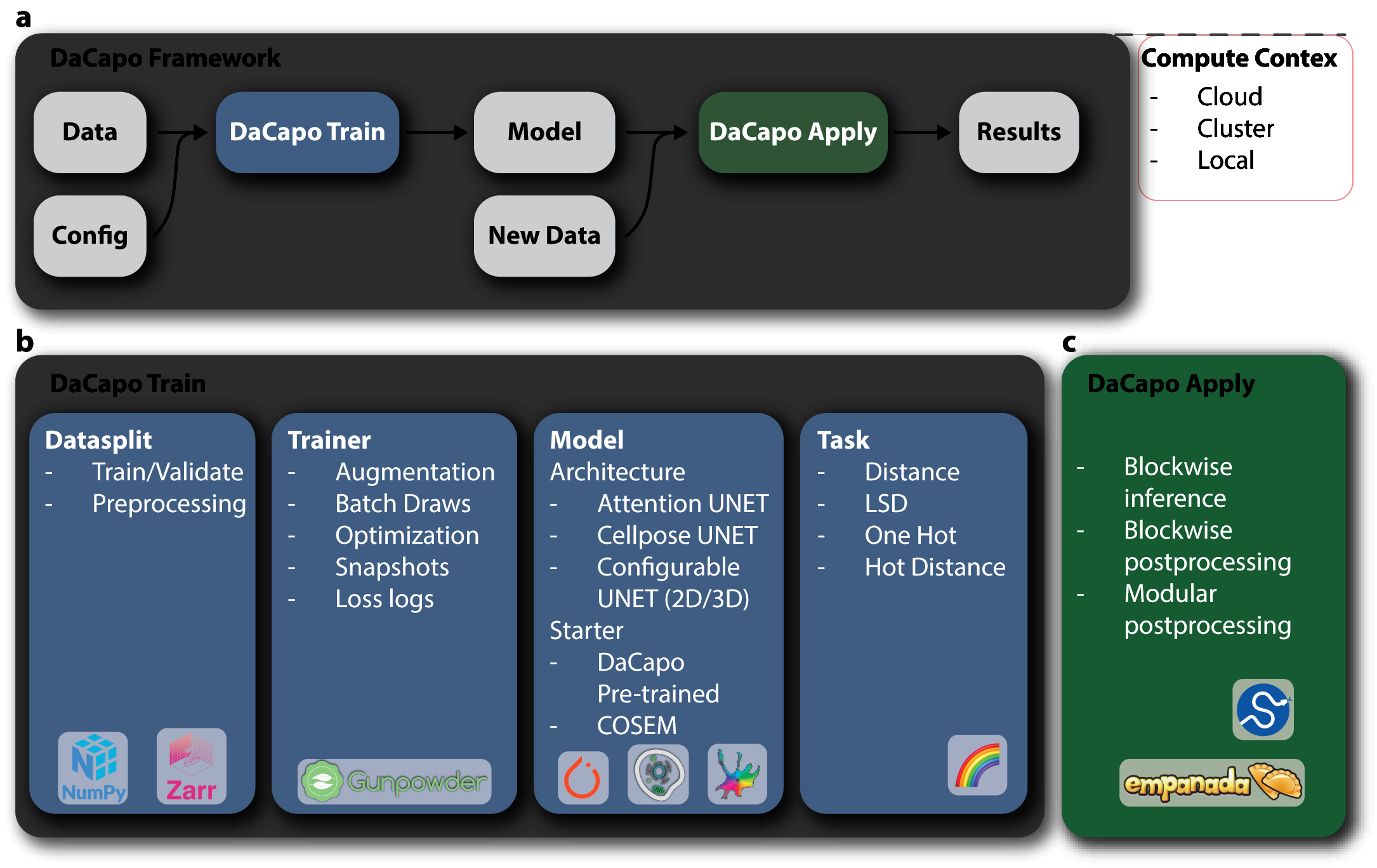
Minimal Tutorial
DaCapo is a framework for easy application of established machine learning techniques on large, multi-dimensional images.
Needed Libraries for this Tutorial
For the tutorial we will use data from the skimage library, and we will use matplotlib to visualize the data. You can install these libraries using the following commands:
pip install 'scikit-image[data]'
pip install matplotlib
Introduction and overview
In this tutorial we will cover the basics of running an ML experiment with DaCapo.
DaCapo has 4 major configurable components:
dacapo.datasplits.DataSplit
dacapo.tasks.Task
dacapo.architectures.Architecture
dacapo.trainers.Trainer
These are then combined in a single dacapo.experiments.Run that includes your starting point (whether you want to start training from scratch or continue off of a previously trained model) and stopping criterion (the number of iterations you want to train).
Environment setup
If you have not already done so, you will need to install DaCapo. You can do this by first creating a new environment and then installing DaCapo using pip.
conda create -n dacapo python=3.10
conda activate dacapo
Then, you can install DaCapo using pip, via GitHub:
pip install git+https://github.com/janelia-cellmap/dacapo.git
pip install dacapo-ml
Be sure to select this environment in your Jupyter notebook or JupyterLab.
Config Store
Configs, model checkpoints, stats, and snapshots can be saved in:
a local folder
an S3 bucket
a MongoDB server
To define where the data goes, create a dacapo.yaml configuration file either in ~/.config/dacapo/dacapo.yaml or in ./dacapo.yaml. Here is a template:
type: files
runs_base_dir: /path/to/my/data/storage
Alternatively, you can define it by setting an environment variable: DACAPO_OPTIONS_FILE=/PATH/TO/MY/DACAPO_FILES.
The runs_base_dir defines where your on-disk data will be stored. The type setting determines the database backend. The default is files, which stores the data in a file tree on disk. Alternatively, you can use mongodb to store the data in a MongoDB database. To use MongoDB, you will need to provide a mongodbhost and mongodbname in the configuration file:
mongodbhost: mongodb://dbuser:dbpass@dburl:dbport/
mongodbname: dacapo
# First we need to create a config store to store our configurations
import multiprocessing
# This line is mostly for MacOS users to avoid a bug in multiprocessing
multiprocessing.set_start_method("fork", force=True)
from dacapo.store.create_store import create_config_store, create_stats_store
config_store = create_config_store()
/opt/hostedtoolcache/Python/3.10.16/x64/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Creating FileConfigStore:
path: /home/runner/dacapo/configs
Data Preparation
DaCapo works with zarr, so we will download skimage example cell data and save it as a zarr file.
import numpy as np
from funlib.geometry import Coordinate, Roi
from funlib.persistence import prepare_ds
from scipy.ndimage import label
from skimage import data
from skimage.filters import gaussian
# Download the data
cell_data = (data.cells3d().transpose((1, 0, 2, 3)) / 256).astype(np.uint8)
# Handle metadata
offset = Coordinate(0, 0, 0)
voxel_size = Coordinate(290, 260, 260)
axis_names = ["c^", "z", "y", "x"]
units = ["nm", "nm", "nm"]
# Create the zarr array with appropriate metadata
cell_array = prepare_ds(
"cells3d.zarr/raw",
cell_data.shape,
offset=offset,
voxel_size=voxel_size,
axis_names=axis_names,
units=units,
mode="w",
dtype=np.uint8,
)
# Save the cell data to the zarr array
cell_array[cell_array.roi] = cell_data
# Generate and save some pseudo ground truth data
mask_array = prepare_ds(
"cells3d.zarr/mask",
cell_data.shape[1:],
offset=offset,
voxel_size=voxel_size,
axis_names=axis_names[1:],
units=units,
mode="w",
dtype=np.uint8,
)
cell_mask = np.clip(gaussian(cell_data[1] / 255.0, sigma=1), 0, 255) * 255 > 30
not_membrane_mask = np.clip(gaussian(cell_data[0] / 255.0, sigma=1), 0, 255) * 255 < 10
mask_array[mask_array.roi] = cell_mask * not_membrane_mask
# Generate labels via connected components
labels_array = prepare_ds(
"cells3d.zarr/labels",
cell_data.shape[1:],
offset=offset,
voxel_size=voxel_size,
axis_names=axis_names[1:],
units=units,
mode="w",
dtype=np.uint8,
)
labels_array[labels_array.roi] = label(mask_array.to_ndarray(mask_array.roi))[0]
print("Data saved to cells3d.zarr")
import zarr
print(zarr.open("cells3d.zarr").tree())
Data saved to cells3d.zarr
/
├── labels (60, 256, 256) uint8
├── mask (60, 256, 256) uint8
└── raw (2, 60, 256, 256) uint8
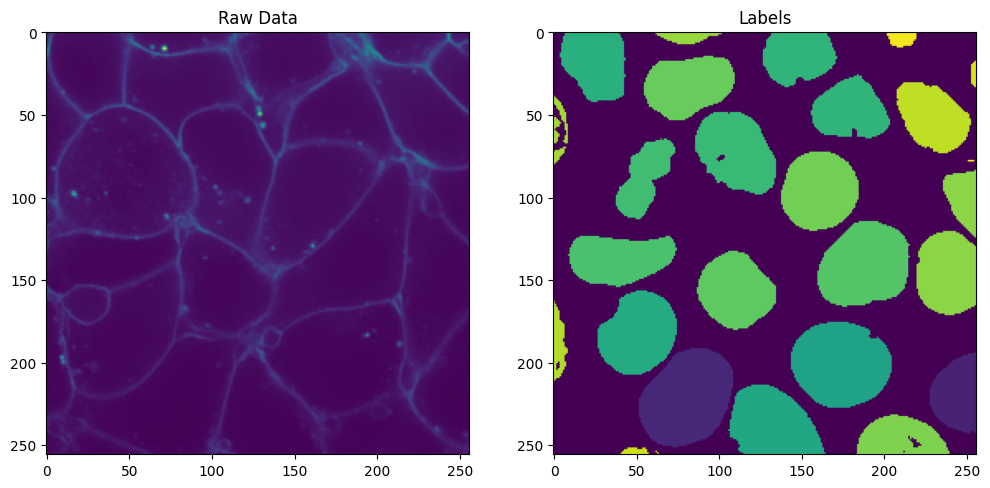
Here we show a slice of the raw data:
# a custom label color map for showing instances
import matplotlib.pyplot as plt
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
# Show the raw data
axes[0].imshow(cell_array.data[0, 30])
axes[0].set_title("Raw Data")
# Show the labels using the custom label color map
axes[1].imshow(labels_array.data[30])
axes[1].set_title("Labels")
plt.show()
Datasplit
Where can you find your data? What format is it in? Does it need to be normalized? What data do you want to use for validation?
We have already saved some data in cells3d.zarr. We will use this data for
training and validation. We only have one dataset, so we will be using the
same data for both training and validation. This is not recommended for real
experiments, but is useful for this tutorial.
from dacapo.experiments.datasplits.simple_config import SimpleDataSplitConfig
from funlib.geometry import Coordinate
datasplit_config = SimpleDataSplitConfig(name="cells3d", path="cells3d.zarr")
datasplit = datasplit_config.datasplit_type(datasplit_config)
config_store.store_datasplit_config(datasplit_config)
datasplit = datasplit_config.datasplit_type(datasplit_config)
# viewer = datasplit._neuroglancer()
config_store.store_datasplit_config(datasplit_config)
Task
What do you want to learn?
Instance Segmentation: Identify and separate individual objects within an image.
Affinities: Learn the likelihood of neighboring pixels belonging to the same object.
Distance Transform: Calculate the distance of each pixel to the nearest object boundary.
Foreground/Background: Distinguish between object pixels and background pixels.
Each of these tasks is commonly learned and evaluated with specific loss functions and evaluation metrics. Some tasks may also require specific non-linearities or output formats from your model.
from dacapo.experiments.tasks import DistanceTaskConfig, AffinitiesTaskConfig
resolution = 260 # nm
# an example distance task configuration
# note that the clip_distance, tol_distance, and scale_factor are in nm
dist_task_config = DistanceTaskConfig(
name="example_dist",
channels=["cell"],
clip_distance=resolution * 10.0,
tol_distance=resolution * 10.0,
scale_factor=resolution * 20.0,
)
# if the config already exists, delete it first
# config_store.delete_task_config(dist_task_config.name)
config_store.store_task_config(dist_task_config)
# an example affinities task configuration
affs_task_config = AffinitiesTaskConfig(
name="example_affs",
neighborhood=[(1, 0, 0), (0, 1, 0), (0, 0, 1)],
)
# config_store.delete_task_config(dist_task_config.name)
config_store.store_task_config(affs_task_config)
Architecture
The setup of the network you will train. Biomedical image to image translation often utilizes a UNet, but even after choosing a UNet you still need to provide some additional parameters. How much do you want to downsample? How many convolutional layers do you want?
from dacapo.experiments.architectures import CNNectomeUNetConfig
# Note we make this UNet 2D by defining kernel_size_down, kernel_size_up, and downsample_factors
# all with 1s in z meaning no downsampling or convolving in the z direction.
architecture_config = CNNectomeUNetConfig(
name="example_unet",
input_shape=(2, 132, 132),
eval_shape_increase=(8, 32, 32),
fmaps_in=2,
num_fmaps=8,
fmaps_out=8,
fmap_inc_factor=2,
downsample_factors=[(1, 4, 4), (1, 4, 4)],
kernel_size_down=[[(1, 3, 3)] * 2] * 3,
kernel_size_up=[[(1, 3, 3)] * 2] * 2,
constant_upsample=True,
padding="valid",
)
config_store.store_architecture_config(architecture_config)
Trainer
How do you want to train? This config defines the training loop and how the other three components work together. What sort of augmentations to apply during training, what learning rate and optimizer to use, what batch size to train with.
from dacapo.experiments.trainers import GunpowderTrainerConfig
trainer_config = GunpowderTrainerConfig(
name="example",
batch_size=10,
learning_rate=0.0001,
num_data_fetchers=1,
snapshot_interval=1000,
min_masked=0.05,
clip_raw=False,
)
config_store.store_trainer_config(trainer_config)
Run
Now that we have our components configured, we just need to combine them into a run and start training. We can have multiple repetitions of a single set of configs in order to increase our chances of finding an optimum.
from dacapo.experiments import RunConfig
from dacapo.experiments.run import Run
iterations = 2000
validation_interval = iterations // 4
run_config = RunConfig(
name="example_run",
datasplit_config=datasplit_config,
task_config=affs_task_config,
architecture_config=architecture_config,
trainer_config=trainer_config,
num_iterations=iterations,
validation_interval=validation_interval,
repetition=0,
)
config_store.store_run_config(run_config)
Retrieve Configurations
All of the configurations are saved in the config store. You can retrieve them as follows:
Architectures: These define the network architectures used in your experiments.
architectures = config_store.retrieve_architecture_configs()
Tasks: These specify the tasks that your model will learn, such as instance segmentation or affinity prediction.
tasks = config_store.retrieve_task_configs()
Trainers: These configurations define how the training process is conducted, including parameters like batch size and learning rate.
trainers = config_store.retrieve_trainer_configs()
Datasplits: These configurations specify how your data is split into training, validation, and test sets.
datasplits = config_store.retrieve_datasplit_configs()
Runs: These combine all the above configurations into a single experiment run.
runs = config_store.retrieve_run_configs()
Train
NOTE: The run stats are stored in the runs_base_dir/stats directory.
You can delete this directory to remove all stored stats if you want to re-run training.
Otherwise, the stats will be appended to the existing files, and the run won’t start
from scratch. This may cause errors.
from dacapo.train import train_run
# from dacapo.validate import validate
from dacapo.experiments.run import Run
from dacapo.store.create_store import create_config_store
config_store = create_config_store()
run = Run(config_store.retrieve_run_config("example_run"))
if __name__ == "__main__":
train_run(run)
Creating FileConfigStore:
path: /home/runner/dacapo/configs
Starting/resuming training for run example_run...
Creating FileStatsStore:
path : /home/runner/dacapo/stats
Trained until 2000, but validated until 2001! Deleting extra validation stats
Current state: trained until 2000/2000
Creating local weights store in directory /home/runner/dacapo
Resuming training from iteration 2000
Retrieving weights for run example_run, iteration 2000
Trained until 2000. Finished.
Visualize
Let’s visualize the results of the training run. DaCapo saves a few artifacts during training including snapshots, validation results, and the loss.
stats_store = create_stats_store()
training_stats = stats_store.retrieve_training_stats(run_config.name)
stats = training_stats.to_xarray()
print(stats)
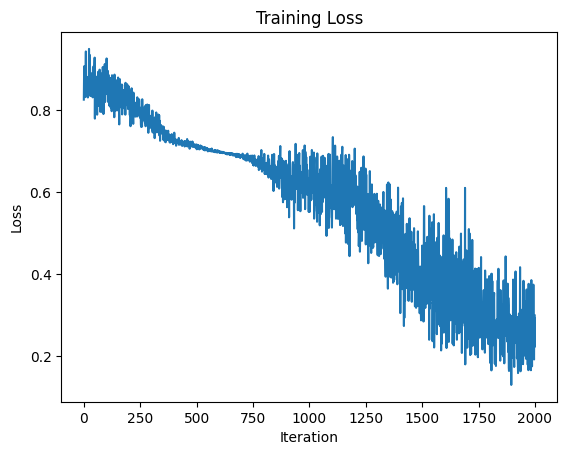
plt.plot(stats)
plt.title("Training Loss")
plt.xlabel("Iteration")
plt.ylabel("Loss")
plt.show()
Creating FileStatsStore:
path : /home/runner/dacapo/stats
<xarray.DataArray (iterations: 2000)> Size: 16kB
array([0.8252877 , 0.88054013, 0.90699542, ..., 0.29377544, 0.22179246,
0.29911986])
Coordinates:
* iterations (iterations) int64 16kB 0 1 2 3 4 5 ... 1995 1996 1997 1998 1999
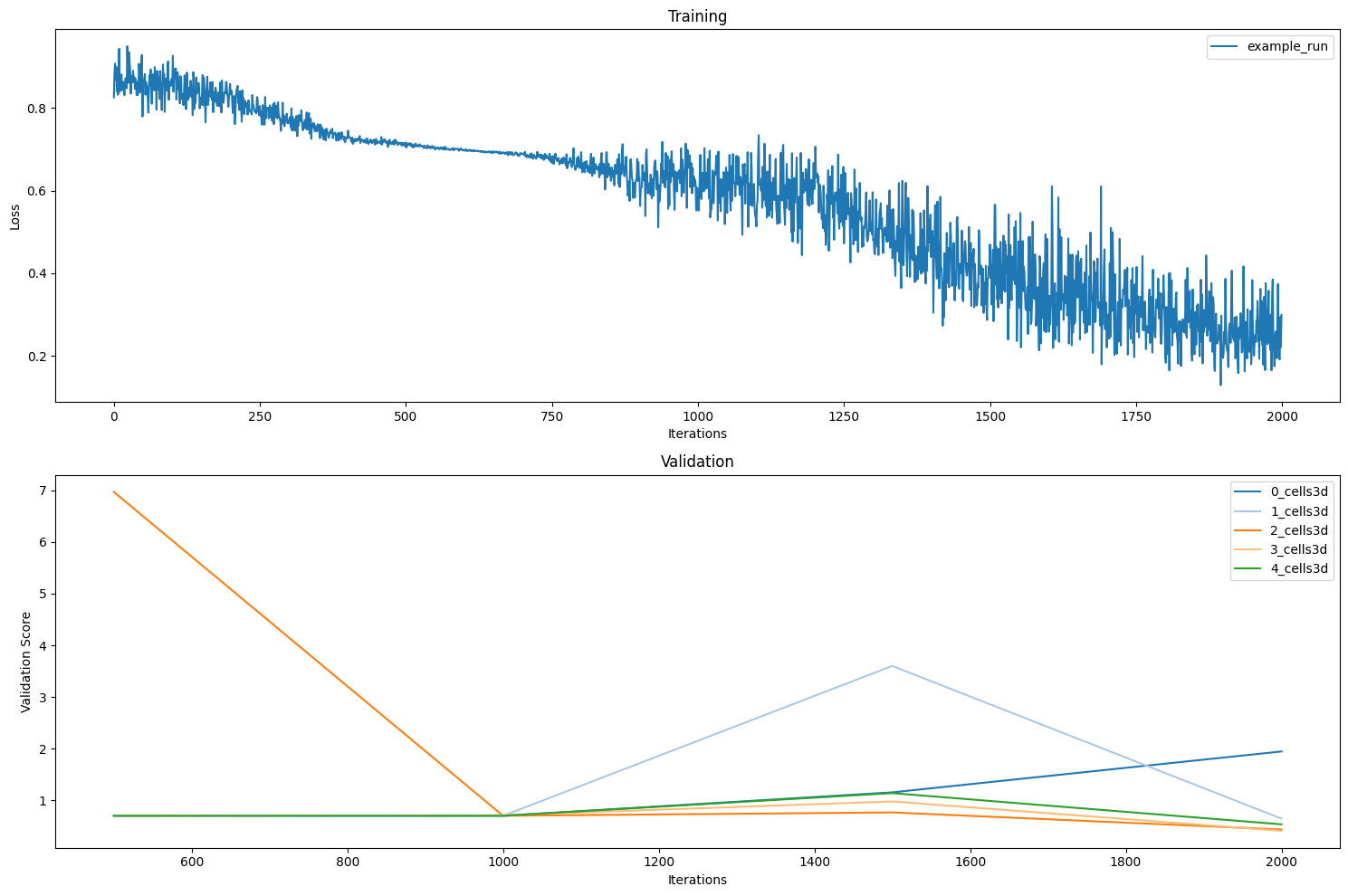
from dacapo.plot import plot_runs
plot_runs(
run_config_base_names=[run_config.name],
validation_scores=["voi"],
plot_losses=[True],
)
# # other ways to visualize the training stats
# stats_store = create_stats_store()
# training_stats = stats_store.retrieve_training_stats(run_config.name)
# stats = training_stats.to_xarray()
# plt.plot(stats)
# plt.title("Training Loss")
# plt.xlabel("Iteration")
# plt.ylabel("Loss")
# plt.show()
Creating FileConfigStore:
path: /home/runner/dacapo/configs
Creating FileStatsStore:
path : /home/runner/dacapo/stats
import zarr
from matplotlib.colors import ListedColormap
np.random.seed(1)
colors = [[0, 0, 0]] + [list(np.random.choice(range(256), size=3)) for _ in range(254)]
label_cmap = ListedColormap(colors)
run_path = config_store.path.parent / run_config.name
# BROWSER = False
num_snapshots = run_config.num_iterations // run_config.trainer_config.snapshot_interval
if num_snapshots > 0:
fig, ax = plt.subplots(num_snapshots, 3, figsize=(10, 2 * num_snapshots))
# Set column titles
column_titles = ["Raw", "Target", "Prediction"]
for col in range(3):
ax[0, col].set_title(column_titles[col])
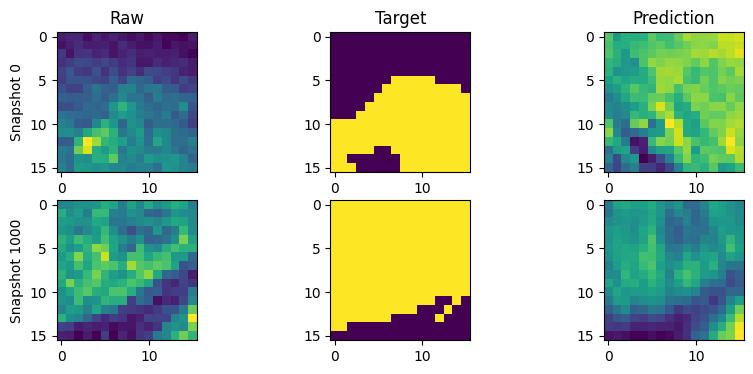
for snapshot in range(num_snapshots):
snapshot_it = snapshot * run_config.trainer_config.snapshot_interval
# break
raw = zarr.open(f"{run_path}/snapshot.zarr/{snapshot_it}/volumes/raw")[:]
target = zarr.open(f"{run_path}/snapshot.zarr/{snapshot_it}/volumes/target")[0]
prediction = zarr.open(
f"{run_path}/snapshot.zarr/{snapshot_it}/volumes/prediction"
)[0]
c = (raw.shape[2] - target.shape[1]) // 2
ax[snapshot, 0].imshow(raw[1, raw.shape[0] // 2, c:-c, c:-c])
ax[snapshot, 1].imshow(target[target.shape[0] // 2])
ax[snapshot, 2].imshow(prediction[prediction.shape[0] // 2])
ax[snapshot, 0].set_ylabel(f"Snapshot {snapshot_it}")
plt.show()
# # %%
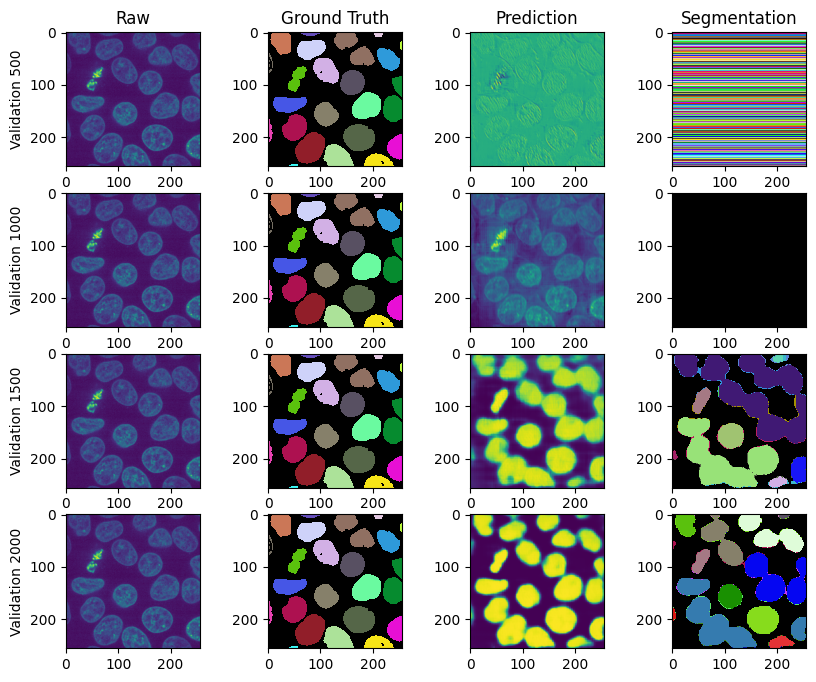
# Visualize validations
import zarr
num_validations = run_config.num_iterations // run_config.validation_interval
fig, ax = plt.subplots(num_validations, 4, figsize=(10, 2 * num_validations))
# Set column titles
column_titles = ["Raw", "Ground Truth", "Prediction", "Segmentation"]
for col in range(len(column_titles)):
ax[0, col].set_title(column_titles[col])
for validation in range(1, num_validations + 1):
dataset = run.datasplit.validate[0].name
validation_it = validation * run_config.validation_interval
# break
raw = zarr.open(f"{run_path}/validation.zarr/inputs/{dataset}/raw")
gt = zarr.open(f"{run_path}/validation.zarr/inputs/{dataset}/gt")
pred_path = f"{run_path}/validation.zarr/{validation_it}/{dataset}/prediction"
out_path = f"{run_path}/validation.zarr/{validation_it}/{dataset}/output/WatershedPostProcessorParameters(id=2, bias=0.5, context=(32, 32, 32))"
output = zarr.open(out_path)[:]
prediction = zarr.open(pred_path)[0]
c = (raw.shape[2] - gt.shape[1]) // 2
if c != 0:
raw = raw[:, :, c:-c, c:-c]
ax[validation - 1, 0].imshow(raw[1, raw.shape[1] // 2])
ax[validation - 1, 1].imshow(
gt[gt.shape[0] // 2], cmap=label_cmap, interpolation="none"
)
ax[validation - 1, 2].imshow(prediction[prediction.shape[0] // 2])
ax[validation - 1, 3].imshow(
output[output.shape[0] // 2], cmap=label_cmap, interpolation="none"
)
ax[validation - 1, 0].set_ylabel(f"Validation {validation_it}")
plt.show()